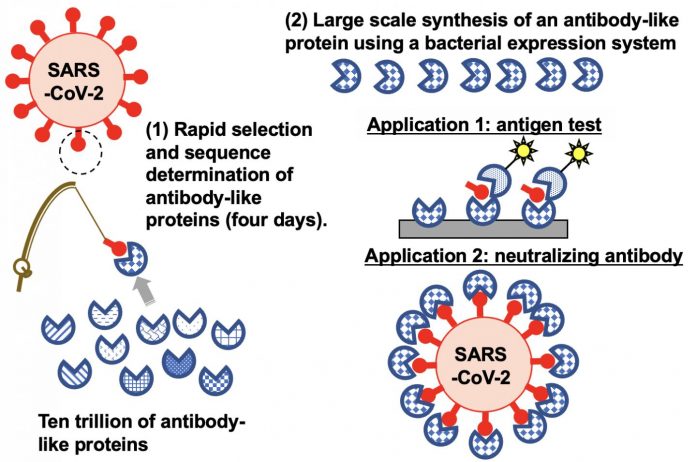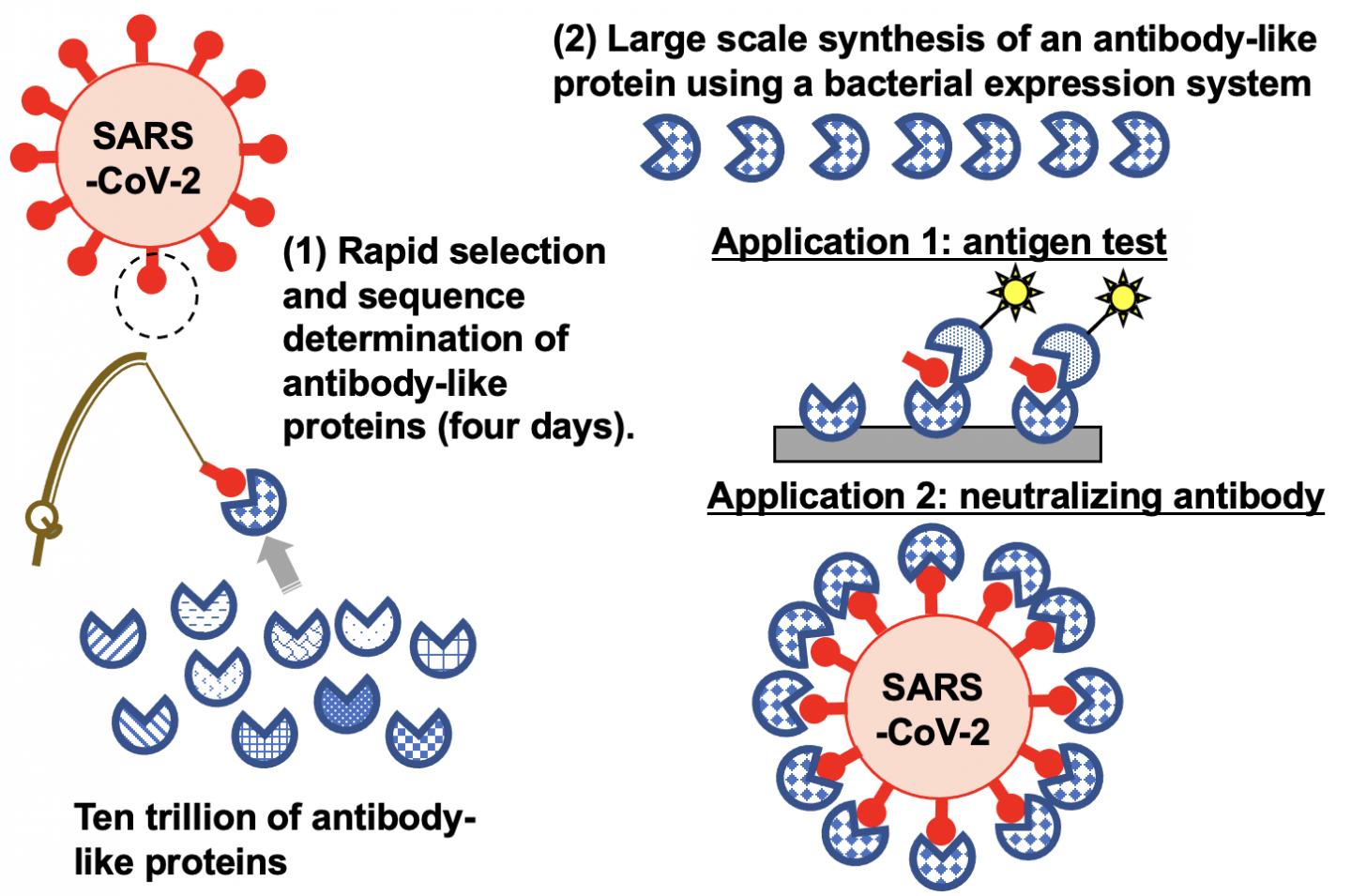
Photo: The TRAP display method “fishes ” for synthetic proteins from among a library of trillions for those that can target SARS-CoV-2. The approach was able to identify proteins that can be…
view more
Credit Image: Hiroshi Murakami
A research team led by Nagoya University scientists in Japan has developed an approach that can quickly find synthetic proteins that specifically bind to important targets, such as components of the SARS-CoV-2 virus. The method was published in the journal Science Advances and could be used to develop test kits or for finding treatments.
“We developed a laboratory technique for rapid selection of synthetic proteins that strongly bind to SARS-CoV-2,” says Nagoya University biomolecular engineer Hiroshi Murakami. “High-affinity synthetic proteins can be used to develop sensitive antigen tests for SARS-CoV-2 and for future use as neutralization antibodies in infected patients.”
Murakami and his colleagues had previously developed a protein selection lab test called TRAP display, which stands for ‘transcription-translation coupled with association of puromycin linker.’ Their approach skips two time-consuming steps in another commonly used technique for searching through synthetic protein libraries. But their investigations indicated there was a problem with the puromycin linker.
In the current study, the team improved their technique by modifying the puromycin linker. Ultimately, they were able to use their TRAP display to identify nine synthetic proteins that bind to the spike protein on SARS-CoV-2’s outer membrane. The approach took only four days compared to the weeks it would take using the commonly used messenger RNA display technology.
TRAP display involves using a large number of DNA templates that code for and synthesize trillions of proteins carrying random peptide sequences. The synthetic proteins are linked to DNA with the help of the modified puromycin linker and then exposed to a target protein. When the whole sample is washed, only the synthetic proteins that bind to the target remain. These are then placed back into the TRAP display for further rounds until only a small number of very specific target-binding synthetic proteins are left.
The researchers investigated the nine synthetic proteins that were found to bind to SARS-CoV-2. Some were specifically able to detect SARS-CoV-2 in nasal swabs from COVID-19 patients, indicating they could be used in test kits. One also attaches to the virus to prevent it from binding to the receptors it uses to gain access to human cells. This suggests this protein could be used as a treatment strategy.
“Our high-speed, improved TRAP display could be useful for implementing rapid responses to subspecies of SARS-CoV-2 and to other potential new viruses causing future pandemics,” says Murakami.
###
This study, “Antibody-like proteins that capture and neutralize SARS-CoV-2,” was published online in Science Advances on September 18, 2020 at doi:10.1126/sciadv.abd3916.
About Nagoya University, Japan
Nagoya University has a history of about 150 years, with its roots in a temporary medical school and hospital established in 1871, and was formally instituted as the last Imperial University of Japan in 1939. Although modest in size compared to the largest universities in Japan, Nagoya University has been pursuing excellence since its founding. Six of the 18 Japanese Nobel Prize-winners since 2000 did all or part of their Nobel Prize-winning work at Nagoya University: four in Physics – Toshihide Maskawa and Makoto Kobayashi in 2008, and Isamu Akasaki and Hiroshi Amano in 2014; and two in Chemistry – Ryoji Noyori in 2001 and Osamu Shimomura in 2008. In mathematics, Shigefumi Mori did his Fields Medal-winning work at the University. A number of other important discoveries have also been made at the University, including the Okazaki DNA Fragments by Reiji and Tsuneko Okazaki in the 1960s; and depletion forces by Sho Asakura and Fumio Oosawa in 1954.
Website: http://en.
TDnews (tunisiesoir.com)