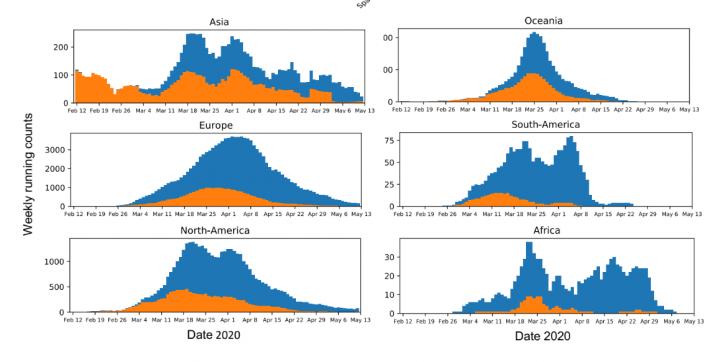
IMAGE: This figure shows the running weekly average counts of sampled SARS-CoV-2 sequences exhibiting the D variant (orange) and G variant (blue) in different continents between January 12 and May 12….
view more
Credit: Korber et al. / Cell
On May 5, 2020, news broke about a reportedly more contagious variant of SARS-CoV-2–the virus that causes COVID-19–based on a preliminary paper posted to the preprint server bioRxiv. The preprint stated that a variant of the virus with a particular mutation leading to an amino acid change, D614G, in its spike protein was “more transmissible” than other forms and represented an “urgent concern” for containment and vaccine development. But in the days that followed, criticisms of these assertions surfaced. On July 2, the journal Cell published a revised and peer-reviewed version of the paper that offers additional experimental and clinical data about the D614G variant suggesting that it may be more infectious, but concludes that we still cannot be certain about whether the variant makes SARS-CoV-2 more transmissible or leads to more severe disease.
“We could see at the time of our initial preprint submission that the G614 variant was becoming the predominant form globally, but we could not differentiate between three broad possibilities that might explain a fitness advantage,” says lead author Bette Korber, a laboratory fellow at Los Alamos National Laboratory whose research prior to her work on COVID-19 focused on the search for an HIV vaccine. “The added experiments in the published study point to enhanced infectivity due to the spike protein change as the favored hypothesis. But infectiousness and transmissibility are not always synonymous, and we hope others will study these viruses in greater detail with wild-type virus in natural infection settings and varied target cells.”
“The Korber et al. paper has changed pretty considerably from what I saw in their preprint,” says Nathan Grubaugh, a virologist at the Yale School of Public Health not affiliated with Korber’s team and the lead author of a Preview contextualizing the paper, also published in Cell. “The in vitro data strengthened the clinical findings, both of which suggest that viruses containing the D614G mutation may be able to replicate to higher levels in human cells. But what we cannot say is that it is more transmissible or leads to more severe disease. Essentially, we don’t know if this has had any meaningful impact on the COVID-19 pandemic.”
While coronaviruses generally have low rates of mutation, Korber and her colleagues were concerned that even small mutations to SARS-CoV-2 could hinder efforts to understand and fight the virus. “We knew from our direct experience in the HIV field that in some cases, a single amino acid change can have a major phenotypic impact,” she says. To that end, the team worked to develop a publicly available data-analysis pipeline that could mine SARS-CoV-2 sequences made available on the Global Initiative for Sharing All Influenza Data (GISAID) database to help scientists explore potentially interesting mutations. They quickly identified the D614G variant as something to pay attention to: its key amino acid change from aspartic acid (D) to glycine (G) occurred on a protein that’s crucial to how the virus infects human cells–and it was rapidly becoming the dominant version of the virus around the world.
The preprint of the paper focused on the development of this tool and offered an analysis of the global prevalence of the G variant of the virus. This analysis suggested that the G variant took over nearly everywhere it was introduced, which the team argued meant that it was outcompeting the D variant: it was better at jumping from human to human. Criticisms of the preprint argued that this conclusion overstated the results of the analysis, that the preprint lacked experimental evidence to show that the G variant was better at infecting human cells, and that the authors had ignored other possible explanations for its spread, such as the founder effect, where a mutation happens to land in an environment more suited to it becoming the dominant form.
To address these concerns and those of the peer reviewers, the researchers further segmented their geographic analysis in order to look at changes in the frequency of the D and G variants in all regions at country, state or province, and county or city levels. They added the dates of stay-at-home orders in various regions to their analysis to show that the G variant often continued to take over a region even after travel became restricted, limiting the possibility that it was simply being repeatedly imported. There was also more data available: approximately 30,000 global SAR-CoV-2 sequences to work with when the paper underwent revision after peer review as opposed to approximately 6,000 at the time the preprint was submitted. “The richer dataset supported our original observations and gave us more confidence in the results,” Korber says. “We now show that in almost every case, G614 increased. There were very few exceptions to this pattern, and we characterize two of them, Iceland and Santa Clara county in California, in detail in the paper.”
The researchers were also able to obtain additional clinical data (they looked at 999 patients from the United Kingdom as compared to 470 in the preprint) to show that patients infected with the G variant of the virus had higher levels of viral RNA, which is sometimes correlated with a higher viral load in the body. There was no difference in hospitalization outcomes for patients with one variant versus the other.
Perhaps most importantly, the revised paper now contains the results of two independently conducted sets of experimental studies to assess the infectivity of the G variant based out of the labs of Erica Ollmann Saphire at La Jolla Institute for Immunology and David Montefiori at Duke University. The researchers engineered versions of the virus with the glycine amino acid substitution and then tested how effectively they could infect human cells in a dish. “Virus particles containing the G form of spike on their surface were approximately 3-6 times more infectious,” says Montefiori. “Because the only difference between the two sets of virus particles was D versus G at position 614, the increased infectivity can be directly attributed to the D614G mutation.” He does note that there are limitations to these findings: the researchers weren’t able to use wild-type viruses and did not study the respiratory system cells that SARS-CoV-2 naturally target. There are also other factors involved in real-life transmission of a virus from person to person that may not be accounted for. Despite the limitations, he says the findings are exciting because “they provide a possible biological explanation for the rapid spread of the G form of the virus across the globe.”
“It seems likely that it’s a fitter virus,” agrees Saphire. Her lab was also able to show that antibodies from six people in the San Diego area who had already recovered from COVID-19 were just as effective at neutralizing both the D and G variants. She notes that the San Diegans were infected at a time when both D and G viruses circulated, so the team can’t be sure which type the people were infected with, but their findings still show that a higher concentration of antibodies isn’t needed to neutralize the new, apparently “fitter” variant despite the higher levels of viral RNA it produced. “That’s good news,” she says. “Human convalescent sera can neutralize the new virus just as well or perhaps just a bit better.” This result suggests that for this particular variant, treatments and vaccines already in development–which are overwhelmingly focused on the spike protein and often based on the original version of the virus sequenced in Wuhan–could still be just as effective.
Korber and her colleagues are still glad they posted the preprint when they did. “We carefully weighed our options,” Korber says. “These experimental assays were not easy to develop, and it seemed they were weeks or possibly months away at the time we published the preprint–this assessment turned out to be correct. It seemed important to get the G614 variant immediately into the queue for further study, and we feel our preprint accelerated efforts to enable comparisons of the D614 and G614 spike variants. We and others have now resolved that there is an apparent difference in infectivity between the two variants, and we were able to join in a new collaborative effort to this end that was enabled in part by the preprint.”
“Computational analysis of sequence changes is always faster than wet lab experimentation,” Saphire says. “Although coronaviruses have some proofreading capacity, mutations can emerge, and vigilance, surveillance, and continued study of the virus will be key to ensuring that the drugs, antibodies, and other interventions under development remain effective. This is the reason the pipeline was set up: to detect mutations that could be important in time to make the needed reagents, grow the necessary viruses, and do the experiments to understand if there is an effect.”
“The global expansion of G614, whether through natural selection or chance, means that this variant now is the pandemic,” notes Grubaugh in his Preview. Still, he argues that for the general public, these results don’t really change much. “Mutation and evolution are natural parts of pandemics and viruses being viruses,” he says. “Some of these can slightly change how a virus ‘behaves,’ but they are not switches that can make a virus suddenly an existential threat.
“While there are still important studies needed to determine if this will influence drug or vaccine development in any meaningful way, we don’t expect that D614G will alter our control measures or make individual infections worse,” he adds. “It’s more of a live look into science unfolding: an interesting discovery was made that potentially touches millions of people, but we don’t yet know the full scope or impact. We only just learned about this virus about six months ago, and we’ll learn a lot more in the next six months.”
###
Funding information and acknowledgments are available in the paper.
Cell, Korber et al.: “Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus” https://www.cell.com/cell/fulltext/S0092-8674(20)30820-5
Cell (@CellCellPress), the flagship journal of Cell Press, is a bimonthly journal that publishes findings of unusual significance in any area of experimental biology, including but not limited to cell biology, molecular biology, neuroscience, immunology, virology and microbiology, cancer, human genetics, systems biology, signaling, and disease mechanisms and therapeutics. Visit: http://www.














